| imversion | Display version information |
This command displays image library version information. Last modification date of an image library command can be retrieved using -version parameter.
| imcreate | Create a new image |
| -n <name> | image name in NDA-namespace |
| -t <type> | image type [1=binary |
| -d <depth> | image depth (max. 32 bits) |
| -w <width> | image width in pixels |
| -h <height> | image height in pixels |
| [-ti <index>] | time index for animated images (default: 0) |
| [-ci <index>] | color index for spectral images (default: 0) |
| [-li <index>] | layer index for 3D images (default: 0) |
This command creates a new image with selected type, size, and tags. An image can represent a (x,y)-plane of pixels (regular images) or (spectrum,angle)-plane (Fourier transformed images). Images can be combined to imagelists. Time index is used to define the position of the image in an animation. Color index is used to distinguish images taken from different areas of the color spectrum. Layer index is used for creating 3D voxel representations where images are stacked in piles.
| imgettime | Get the time value from an image |
| -n <name> | image name |
| -v <value> | field name receiving the time value |
| imputtime | Put a time value to an image |
| -n <name> | image name |
| -v <value> | new value |
| imgetcolor | Get the color value from an image |
| -n <name> | image name |
| -v <value> | field name receiving the color value |
| imputcolor | Put a color value to an image |
| -n <name> | image name |
| -v <value> | new value |
| imgetlayer | Get the layer value from an image |
| -n <name> | image name |
| -v <value> | field name receiving the layer value |
| imputlayer | Put a layer value to an image |
| -n <name> | image name |
| -v <value> | new value |
These commands provide a way to read and write image tags. For more information on tags please refer to imcreate above.
| imgetpixel | Get a pixel value from an image |
| -n <name> | image name |
| -x <position> | horizontal pixel index |
| -y <position> | vertical pixel index |
| -v <name> | field name receiving the pixel value |
| imputpixel | Put a pixel value to an image |
| -n <name> | image name |
| -x <position> | horizontal pixel index |
| -y <position> | vertical pixel index |
| -v <value> | new value |
| imgetipixel | Get an imaginary pixel value from an image |
| -n <name> | image name |
| -x <position> | horizontal pixel index |
| -y <position> | vertical pixel index |
| -v <name> | field name receiving the pixel imaginary value |
| imputipixel | Put an imaginary pixel value to an image |
| -n <name> | image name |
| -x <position> | horizontal pixel index |
| -y <position> | vertical pixel index |
| -v <value> | new imaginary value |
These commands offer a low-level access to image's bitplane. They offer only moderate performance and should not be used for laborious tasks. The imaginary versions of the commands are reserved for images where the pixels are complex numbers.
| imconvert | Convert image to a new one |
| -src <name> | source name |
| -dest <name> | destination name |
| -t <type> | image type [1=binary |
| -d <depth> | image depth (max. 32 bits) |
This command converts an existing image to a new one, which is of different type.
| imsize | Get the size of an image |
| -n <name> | image name |
| -fout <size> | field name receiving the image size |
This command inquires the pixel width and height of an image.
| imcopy | Copy an image to another image |
| -src <name> | source name |
| -dest <name> | destination name |
This command allows the user to make copies of images.
| imload | Load an image from a file |
| -n <name> | image name |
| -f <filename> | filename (GIF-file) |
| [-ti <index>] | time index for animated images (default: 0) |
| [-ci <index>] | color index for spectral images (default: 0) |
| [-li <index>] | layer index for 3D images (default: 0) |
| imsave | Save an image into a file |
| -n <name> | image name |
| -f <filename> | filename (GIF-file) |
These commands are used to load/save images from/to the filesystem. The only supported file format is CompuServe Graphics Interchange Format (*.gif).
Example: In this example we load and save an image from/to a file.
imload -n image -f paper.gif -ti 1 -ci 2 -li 0 imsave -n image -f img2.gif
| image2im | Convert an old image format to the new format |
| -src <name> | source name |
| -dest <name> | destination name |
| im2image | Convert an image from the new format to the old format |
| [ | |
| -gray <name> | source name for gray channel |
| -red <name> | source name for red channel |
| -green <name> | source name for green channel |
| -blue <name> | source name for blue channel |
| ] | |
| -dest <name> | destination name |
These commands convert images from/to the old image format which is only required for visualization purposes.
Example: In this example we convert the image formats back and forth.
imload -n image -f paper.gif im2image -gray image -dest oldimage mkgrp g bgimg g -img oldimage show g image2im -src oldimage -dest image2
| imhistogram | Builds a grayscale histogram |
| -src <name> | source name |
| -dest <name> | destination name |
| [-bits <name>] | histogram color model bit count (default is 8) |
This command builds a histogram of the amounts of pixels of each gray intensity level in the image. Full-size 32-bit histogram has ![]() pins, which makes it impractical. Setting parameter -bits to lower value will generate histograms from
pins, which makes it impractical. Setting parameter -bits to lower value will generate histograms from ![]() color model.
color model.
| imconstraint | Set pixel values lower than lower bound and higher than upper bound to specified values |
| -src <name> | source name |
| -dest <name> | destination name |
| -t <type> | threshold type [1=lower |
| [ | |
| -lb <value> | 24-bit lower threshold |
| -lv <value> | new 24-bit value for pixels below lower threshold |
| ] | |
| [ | |
| -ub <value> | 24-bit upper threshold |
| -uv <value> | new 24-bit value for pixels above upper threshold |
| ] |
This command is used to make pixelwise thresholds directly to the image. Pixels, whose value is below the lower bound, are set to the lower value. Pixels whose intensity value is higher than the upper bound are set to the upper value. Intensity values are given in 24-bit format as NDA cannot store the full 32-bit format in it's namespace. In 24-bit model 0 is the lowest value (black) and 16777216 is the highest value (white).
| imcrop | Crop a region from an image to another image |
| -src <name> | source name |
| -dest <name> | destination name |
| -x1 <position> | horizontal start pixel index |
| -y1 <position> | vertical start pixel index |
| -x2 <position> | horizontal end pixel index |
| -y2 <position> | vertical end pixel index |
This command picks a subregion from an image to a new image.
| imnegate | Negate the pixel values of an image and store them into another image |
| -src <name> | source name |
| -dest <name> | destination name |
This command flips the gray intensities in order to make dark areas appear bright and vice versa.
| imfilter | Filter an image |
| -src <name> | source name |
| -dest <name> | destination name |
| -o <operation> | mask operation [0=average |
| -s <size> | mask size in pixels |
This command applies a smoothing filter to the image.
| imfindthreshold | Find a suitable threshold value from an image |
| -src <name> | source name |
| -m <mode> | operation mode [absolut |
| [-p <value>] | percent threshold value used in percent mode |
| -fout <name> | destination name which receives the 24-bit threshold value |
This command attempts to find the best thresholding value for an image. Intensity values are given in 24-bit format as NDA cannot store the full 32-bit format in it's namespace. In 24-bit model 0 is the lowest value (black) and 16777216 is the highest value (white).
| imthreshold | Threshold an image to create a new binary image |
| -src <name> | source name |
| -dest <name> | destination name |
| -value <value> | 24-bit threshold value |
This command performs the actual image thresholding. The new binary image will have black pixels where the gray image has smaller intensity values than threshold value and white pixels where the gray image has higher intensities than the thresholding value.
Example: In this example we load a new image, perform some filtering and then threshold the result to obtain a new binary image.
imload -n image -f paper.gif
imfilter -src image -dest image2 -o 0 -s 2
imfindthreshold -src image2 -m percent -p 20 -fout th
imthreshold -src image2 -dest image3 -value ${th}
im2image -gray image3 -dest oldimage
mkgrp g
bgimg g -img oldimage
show g
| imconvolution | Perform a convolution between an image and a mask |
| -src <name> | source name |
| -dest <name> | destination name |
| -mask <value> | mask name |
| -px <position> | mask origo horizontal position |
| -py <position> | mask origo vertical position |
This command is used to perform a pixelwise convolution.
| imedge | Find edges from an image |
| -src <name> | source name |
| -dest <name> | destination name |
| -o <operation> | operation [g1=Laplace of Gaussian 1 |
This command is used to find borders in grayscale images. NOTE: The result is not automatically thresholded.
| imfft | Remap image data betwen real space and Fourier space |
| -src <name> | source name |
| -dest <name> | destination name |
| [-inv] | inverse Fourier transform |
| [-lefttop] | padding on left and top |
| [-all] | padding on all sides |
| [-rightbottom] | padding on right and bottom |
This command performs the Fourier transform to the the image.
| imshiftspec | Swap quadrants |
| -src <name> | source name |
| -dest <name> | destination name |
This command swaps diagonal quadrants.
| imfreqvec | Swap quadrants |
| -src <name> | source name |
| -dest <name> | destination name |
| [-angle] | do angular analysis |
| [-radius] | do radial analysis |
This command calculates an angular or radial mass distribution.
| imvisualizefft | Reorganize and rescale a Fourier image to a more illustrative image |
| -src <name> | source name |
| -dest <name> | destination name |
| -contrast <value> | contrast value in [0,32] |
| [-shift] | swap quadrants |
| [-scale] | scale values to [0,255] |
This command prepares the image for visualization. The original Fourier transformed image has too wide intensity range for visualization making them look almost entirely black. To remedy this the intensities are rescaled to logarithmic scale. Both diagonal quadrants are also flipped to archieve more familiar representation.
Example: In this example we use Fourier transform to detect regular patterns in a paper sample.
imload -n image -f paper.gif # FFT imfft -src image -dest fftimage -all imvisualizefft -src fftimage -dest fftvisualize -shift -scale im2image -gray fftvisualize -dest fftoldimage mkgrp FFT bgimg FFT -img fftoldimage show FFT # FFT distributions imshiftspec -src fftimage -dest fftshift imfreqvec -src fftshift -dest fftrad -radius imfreqvec -src fftshift -dest fftangle -angle imfreqvec -src fftimage -dest fftxxr -radius imfreqvec -src fftimage -dest fftxxa -angle mkgrp FFTdistributions dis FFTdistributions all en FFTdistributions ldgr saxis ldgrv FFTdistributions -inx 0 -f fftrad -co red ldgrv FFTdistributions -inx 1 -f fftangle -co blue ldgrv FFTdistributions -inx 2 -f fftxxr -co green ldgrv FFTdistributions -inx 3 -f fftxxa -co cyan show FFTdistributions
| imtranslate | Translates images local coordinate system |
| -A <name> | A's name |
| [-x <value>] | horizontal transfer |
| [-y <value>] | vertical transfer |
| imreflect | Reflects images local coordinate system |
| -A <name> | A's name |
| [-x] | reflect horizontally |
| [-y] | reflect vertically |
These commands are used to alter images local coordinate system. This feature is most useful for images which are used as masks for operations.
| imcomplement | Performs complement operation |
| -A <name> | A's name |
| -dest <name> | destination name |
| imdifference | Performs substraction operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imunion | Performs union operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imintersection | Performs intersection operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imerode | Performs erosion operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imdilate | Performs dilation operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imopen | Performs opening operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imclose | Performs closing operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
These commands perform the most widely used morphological operations for binary images. For more information please see (Gonzalez, Woods?) table 8.3 on pages 544-545.
| imhitmiss | Performs hitmiss operation |
| -A <name> | A's name |
| -B1 <name> | B1's name |
| -B2 <name> | B2's name |
| -dest <name> | destination name |
| [-x1 <value>] | B1's horizontal transfer |
| [-y1 <value>] | B1's vertical transfer |
| [-x2 <value>] | B2's horizontal transfer |
| [-y2 <value>] | B2's vertical transfer |
| imboundary | Performs boundary operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imregionfill | Performs regionfill operation |
| -A <name> | A's name |
| -S <name> | S's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imconnected | Performs connected operation |
| -A <name> | A's name |
| -S <name> | S's name |
| -B <name> | B's name |
| -dest <name> | destination name |
| [-x <value>] | B's horizontal transfer |
| [-y <value>] | B's vertical transfer |
| imskeleton | Performs skeleton operation |
| -A <name> | A's name |
| -B <name> | B's name |
| -dest <name> | destination name |
These commands perform more advanced morphological operations for binary images. For more information please see (Gonzalez, Woods?) table 8.3 on pages 544-545.
| imcooccurence | Calculate a co-occurence matrix image from an image |
| -src <name> | source name |
| -dest <name> | destination name |
| -dx <size> | shift in horizontal direction |
| -dy <size> | shift in vertical direction |
| [-sign] |
This command calculates a co-occurence matrix from the image.
| imcofeatures | Calculate co-occurence features from a co-occurence matrix image |
| -src <name> | source name |
| -dout <name> | destination name |
| [-l <value>] | lambda (default: 1) |
| [-k <value>] | kappa (default: 2) |
| [-con] | calculate contrast feature |
| [-cor] | calculate correlation feature |
| [-ene] | calculate energy feature |
| [-ent] | calculate entropy feature |
| [-idm] | calculate idm feature |
| [-max] | calculate mmax feature |
This command calculates numerical descriptors from the co-occurence matrix.
Example: In this example we calculate a co-occurence matrix and its' features.
imload -n image -f paper.gif
imcooccurence -src image -dest comatrix -dx 5 -dy 5
imcofeatures -src comatrix -dout features -con -cor -ene -ent \
-idm -max
| imsegment | Segment binary image to blobs |
| -src <name> | source name |
| -cout <name> | class information destination name |
| -dout <name> | pixel information destination name |
| -blobindex <boolean> | include blob indexes [false |
| imfloodsegment | Segment gray image to blobs |
| -src <name> | source name |
| -cout <name> | class information destination name |
| -dout <name> | pixel information destination name |
| -blobindex <boolean> | include blob indexes [false |
| impeaksegment | Segment gray image to blobs |
| -src <name> | source name |
| -cout <name> | class information destination name |
| -dout <name> | pixel information destination name |
| -blobindex <boolean> | include blob indexes [false |
These commands are used to segment the image into smaller subregions of more interest. imsegment uses a 4-pixel neighbourhood on the binary image to assess which pixels belong to the same subregion a.k.a. segment, blob, grain. imfloodsegment finds high points in the intensity landscape and fills the hills one by one. impeaksegment finds highest points also but fills all the hills at the same time. All of these methods result in a different type of segmentation. All three methods return a classification and a dataframe with fields x and y for pixel locations. Optionally blob index may be stored in blob field.
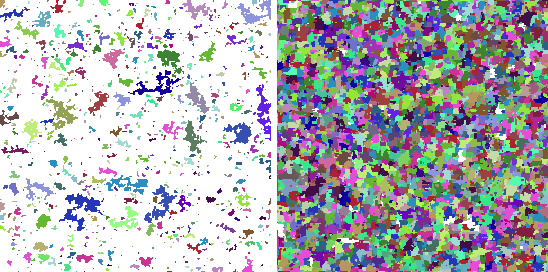

| imsegmentcoloring | Color image segments (blobs) with specified color |
| -src <name> | source name |
| -dest <name> | destination name |
| -segdata <name> | segmentation data |
| -inst <name> | instructions for coloring |
| imfastsegmentcoloring | Color image segments (blobs) with specified color |
| -src <name> | source name |
| -dest <name> | destination name |
| -segcl <name> | segmentation cldata |
| -segdata <name> | segmentation data |
| -inst <name> | instructions for coloring |
These commands allow user to alter segment's pixel values to match a spesific color. The -inst dataframe must contain fields blob which identifies which segment is to be colored and color which has the 0-255 intensity value.
Example: In this example segment the image into small blobs and color some of these blobs.
imload -n image -f paper.gif
imfilter -src image -dest image2 -o 0 -s 2
imfindthreshold -src image2 -m percent -p 20 -fout th
imthreshold -src image2 -dest image3 -value ${th}
imsegment -src image2 -cout segcl -dout segdata -blobindex true
len -n segcl -fout len
serie -len ${len} -fout blob
setdata -f color -t int -len ${len} -vals 0=0;
expr -fout color -expr 'color'+IRAND(256);
select inst -f blob color
imsegmentcoloring -src image2 -dest image3 -segdata segdata \
-inst inst
im2image -gray image3 -dest oldimage
mkgrp g
bgimg g -img oldimage
show g
| imtoppoints | Finds top points from an image |
| -i <name> | source image name |
| -dout <name> | point field information |
| -s <size> | turf size (turf has rectangular shape) |
| [-neq] | deny equal value points inside the mask |
This command attempts to find hilltops in the intensity landscape. The resulting dataframe will contain fields x and y for spatial position. Normally points tolerate other points of the same intensity on their turf. Using [-neq] option removes this possibility.
| imtoppoints2 | Finds top points from an image |
| -i <name> | source image name |
| -dout <name> | point field information |
| -s <size> | turf size (turf has circular shape) |
| [-neq] | deny equal value points inside the mask |
This command is an another way to find hilltops in the intensity landscape. The resulting dataframe will contain fields x and y for spatial position. Normally points tolerate other points of the same intensity on their turf. Using [-neq] option removes this possibility.
| imcenterpoints | Find blob center according to indicator function |
| -i <name> | source image name |
| -c <name> | class information |
| -d <name> | pixel information |
| -dout <name> | point field information |
| immasspoints | Find blob center according to mass function |
| -i <name> | source image name |
| -c <name> | class information |
| -d <name> | pixel information |
| -dout <name> | point field information |
| impeakpoints | Find blob center according to peak indicator function |
| -i <name> | source image name |
| -c <name> | class information |
| -d <name> | pixel information |
| -dout <name> | point field information |
These commands calculate a center for each segment. The resulting dataframe will contain fields x and y for spatial position.
| imgundersen | Classify points in a point field according to their location |
| -i <name> | source image name |
| -c <name> | class information |
| -d <name> | pixel information |
| -dout <name> | point field information |
| -m <name> | border mode [0=percent |
| -v <name> | border width |
This command classifies spatial points according to their location into three categories. This classification is placed in a field named frame. Frame value 0 means that the points is to be included into calculations. Value 1 means that point is included in the calculations but is not included in the actual result. Value 2 means that point lies in the forbidden area and is banned from all calculations. For more information, please see page 223 in Stochastic Geometry and its Applications, by Stoyan, Kendall and Mecke.
| immarks | Calculate point mark information from blob information |
| -i <name> | gray image name |
| -bwi <name> | binary image name |
| -c <name> | class information |
| -d <name> | pixel information |
| -dout <name> | point field information |
| [-avgvar] | include blob's average and variance information |
| [-eig] | include blob's principal components and their weights |
| [-card] | include blob's cardinality |
| [-border] | include blob's border's cardinality |
| [-mass] | include blob's mass infomation |
| [-minmax] | include blob's minimum and maximum intensity information |
| [-hist] | include blob's grayscale histogram's information |
| [-rou] | include roundness information |
| [-rad] | include radii information |
| [-ecc] | include eccentricity information |
| [-ste] | include steepness information |
| [-ori] | include orientation information |
This command calculates descriptive features from the segments.
Example: In this example we form a point field based on segmentation information. In order to avoid border effects we use a technique called Gundersen framing. We also count some mark information to describe these points.
imload -n image -f paper.gif
imfilter -src image -dest image2 -o 0 -s 2
imfindthreshold -src image2 -m percent -p 20 -fout th
imthreshold -src image2 -dest image3 -value ${th}
imsegment -src image2 -cout segcl -dout segdata -blobindex true
immasspoints -i image2 -c segcl -d segdata -dout points
imgundersen -i image2 -c segcl -d segdata -dout points -m 1 -v 50
immarks -i image -c segcl -d segdata -dout points \
-card -border -hist
| impointfieldstat | Calculate point field second- and third-order statistics |
| -i <name> | source image name |
| -d <name> | point field information |
| -dout <name> | statistical curves |
| [-fout <name>] | estimated lambda (average intensity) |
| -pnts <count> | point count in curves |
| -radd <count> | r step between points |
| [-px <position>] | horizontal point position for Lx and gx |
| [-py <position>] | vertical point position for Lx and gx |
| [-m <field>] | mark field for mark correlation |
| [-test <test>] | test for mark correlation [1=area |
| [-mask <mask>] | mask type for third-order statistics [1=rectangle |
| [-asize <size>] | mask width for third-order statistics |
| [-bsize <size>] | mask height for third-order statistics |
| [-r] | include r-axis |
| [-K] | include Ripleys K-function |
| [-L] | include L-function |
| [-g] | include pair correlation function |
| [-Lx] | include local L-function |
| [-gx] | include local pair correlation function |
| [-k] | include mark correlation function |
| [-z] | include third-order statistic function |
| [-ref] | include Poisson functions for reference |
This command calculates point field statistics. For more information, please see pages 275-305 in Fractals, Random Shapes and Point Fields, by Stoyan and Stoyan.
Example: In this example we calculate Ripleys K-function of the point field.
impointfieldstat -i image -d points -dout k -pnts 101 \
-radd 0.5 -r -K -ref
mkgrp K
select stat -f k.K k.ref_K
fldstat -d stat -dout stat2 -min -max
fldstat -d stat2 -dout stat3 -min -max
len -fout len -n k.r
expr -fout len -expr 'len'-1;
ldgrv K -inx 0 -f k.ref_K -co red -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]}
ldgrv K -inx 0 -f k.K -co black -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]}
saxis K -inx 0 -ax x -sca abs -min ${k.r[0]} \
-max ${k.r[${len}]} -step 2
saxis K -inx 0 -ax y -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]} -step 2
rm stat
rm stat2
rm stat3
rm len
viewp K -dz -150
show K
Example: In this example we calculate local pair correlation function of the point field.
impointfieldstat -i image2 -d points -dout gx -pnts 101 \
-radd 0.5 -r -gx -ref -px 256 -py 256
mkgrp Gx
select stat -f gx.gx gx.ref_gx
fldstat -d stat -dout stat2 -min -max
fldstat -d stat2 -dout stat3 -min -max
len -fout len -n gx.r
expr -fout len -expr 'len'-1;
ldgrv Gx -inx 0 -f gx.ref_gx -co red -sca abs \
-min ${stat3.min[0]} -max ${stat3.max[1]}
ldgrv Gx -inx 0 -f gx.gx -co black -sca abs \
-min ${stat3.min[0]} -max ${stat3.max[1]}
saxis Gx -inx 0 -ax x -sca abs -min ${gx.r[0]} \
-max ${gx.r[${len}]} -step 2
saxis Gx -inx 0 -ax y -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]} -step 2
rm stat
rm stat2
rm stat3
rm len
viewp Gx -dz -150
show Gx
Example: In this example we calculate mark correlation function of the point field.
impointfieldstat -i image2 -d points -dout markcorrelation \
-pnts 101 -radd 0.5 -r -k -ref -m are -test 1
mkgrp MarkCorrelation
select stat -f markcorrelation.k_are markcorrelation.ref_k_are
fldstat -d stat -dout stat2 -min -max
fldstat -d stat2 -dout stat3 -min -max
len -fout len -n markcorrelation.r
expr -fout len -expr 'len'-1;
ldgrv MarkCorrelation -inx 0 -f markcorrelation.ref_k_are \
-co red -sca abs -min ${stat3.min[0]} -max ${stat3.max[1]}
ldgrv MarkCorrelation -inx 0 -f markcorrelation.k_are -co black \
-sca abs -min ${stat3.min[0]} -max ${stat3.max[1]}
saxis MarkCorrelation -inx 0 -ax x -sca abs \
-min ${markcorrelation.r[0]} \
-max ${markcorrelation.r[${len}]} -step 2
saxis MarkCorrelation -inx 0 -ax y -sca abs \
-min ${stat3.min[0]} -max ${stat3.max[1]} -step 2
rm stat
rm stat2
rm stat3
rm len
viewp MarkCorrelation -dz -150
show MarkCorrelation
Example: In this example we calculate the third-order statistic of the point field.
impointfieldstat -i image2 -d points -dout z -pnts 101 -radd 0.5 \
-r -z -ref -mask 1 -asize 0.5 -bsize 0.25
mkgrp Z
select stat -f z.z z.ref_z
fldstat -d stat -dout stat2 -min -max
fldstat -d stat2 -dout stat3 -min -max
len -fout len -n z.r
expr -fout len -expr 'len'-1;
ldgrv Z -inx 0 -f z.ref_z -co red -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]}
ldgrv Z -inx 0 -f z.z -co black -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]}
saxis Z -inx 0 -ax x -sca abs -min ${z.r[0]} \
-max ${z.r[${len}]} -step 2
saxis Z -inx 0 -ax y -sca abs -min ${stat3.min[0]} \
-max ${stat3.max[1]} -step 2
rm stat
rm stat2
rm stat3
rm len
viewp Z -dz -150
show Z
| imnearestneighbour | Calculate contact information in a point field |
| -d <name> | point field information |
| -dout <name> | point field information |
| -k <count> | count values for 1-k neighbours |
| [-index] | include indexes to nearest neigbours |
| [-angle] | include angles to nearest neigbours |
| [-cumangle] | include cumulative angles to nearest neigbours |
| [-contangle] | include continuing angle to nearest neigbours |
| [-dist] | include distances to nearest neigbours |
| [-cumdist] | include cumulative distances to nearest neigbours |
| [-contdist] | include continuing distances to nearest neigbours |
This command calculates nearest neighbour information.
Example: In this example we inspect contact distributions in the point field.
... imnearestneighbour -d points -dout points -k 3 -dist -angle
| imrandpoints | Generate a random point field with uniform distribution |
| -dout <name> | point field information |
| -pnts <points> | points in the point field |
| -w <width> | width of the point field |
| -h <height> | height of the point field |
This command is used to generate a random point process for testing purposes. Points are generated from a 2D uniform distribution.
| imkernel2d | Kernel estimate a 2D process |
| -p <name> | point process dataframe |
| -t <name> | test process dataframe |
| -dout <name> | estimated angles, distances, and weights |
| -k <value> | kernel type [1=box |
| -V <value> | kernel size |
| -c <value> | cut distance (ignore points further than this) |
| imknn2d | KNN estimate a 2D process |
| -p <name> | point process dataframe |
| -t <name> | test process dataframe |
| -dout <name> | estimated angles, distances, and weights |
| -k <value> | kernel type [1=box |
| -K <value> | kernel point size |
| -c <value> | cut distance (ignore points further than this) |
| imkernel1d | Kernel estimate a weighted dataset |
| -v <name> | field of data |
| -p <name> | field of priories |
| -dout <name> | estimated distribution |
| -pins <value> | number of pins |
| -a <value> | axis minimum |
| -b <value> | axis maximum |
| -k <value> | kernel type [1=box |
| -V <value> | kernel size |
| -c <value> | cut distance (ignore points further than this) |
| imknn1d | KNN estimate a weighted dataset |
| -v <name> | field of data |
| -p <name> | field of priories |
| -dout <name> | estimated distribution |
| -pins <value> | number of pins |
| -a <value> | axis minimum |
| -b <value> | axis maximum |
| -k <value> | kernel type [1=box |
| -K <value> | kernel point size |
| -c <value> | cut distance (ignore points further than this) |
These commands are used to estimate distributions of the point processes. 2D methods take a point field and return information on all available point pairs. 1D methods are used to generate the distribution based on weighted point pairs.
Example: In this example we use two estimators to estimate the angle and distance distributions from the point process.
...
imrandpoints -dout test -pnts 1000 -w 512 -h 512
imkernel2d -p points -t test -dout distdata1 -V 50
expr -fout distdata1.weight -expr 'distdata1.weight' * 1000000.0;
imkernel1d -v distdata1.angle -p distdata1.weight \
-dout angle1 -V 0.1
imkernel1d -v distdata1.distance -p distdata1.weight \
-dout distance1 -V 1 -a 0 -b 100
mkgrp Angle1
ldgrv Angle1 -f angle1.value
show Angle1
mkgrp Distance1
ldgrv Distance1 -f distance1.value
show Distance1
imknn2d -p points -t test -dout distdata2 -K 10
expr -fout distdata2.weight -expr 'distdata2.weight' * 1000000.0;
imkernel1d -v distdata2.angle -p distdata2.weight \
-dout angle2 -V 0.1
imkernel1d -v distdata2.distance -p distdata2.weight \
-dout distance2 -V 1 -a 0 -b 100
mkgrp Angle2
ldgrv Angle2 -f angle2.value
show Angle2
mkgrp Distance2
ldgrv Distance2 -f distance2.value
show Distance2
| imsimspectralmethod | Simulate an image |
| -iout <name> | image name |
| -w <value> | image width |
| -h <value> | image height |
| -m <method> | image simulation method [ circular |
| -a <value> | image simulation scaling |
This command simulates an image using the spectral method. For more information, please see pages 191-192 in Geostatistical Simulation by C. Lantuejoul.
| imvoronoi | Generates Delaunay/Voronoi diagrams (based on Steven Fortune's algorithm) |
| -d <name> | vertex information |
| -dout <name> | edge information |
| [-dout2 <name>] | point field information (needed for Voronoi diagrams) |
| [-sites <value>] | number of sites |
| [-xmin <value>] | minimum x value |
| [-xmax <value>] | maximum x value |
| [-ymin <value>] | minimum y value |
| [-ymax <value>] | maximum x value |
| [-d] | echo debug information |
| [-s] | data is sorted |
| [-t] | generate Delaunay diagram (default: Voronoi diagram) |
| [-p] | plot information |
This command is used to build Delaunay and Voronoi triangulations based on x and y fields in the vertex dataframe. In Delaunay case, a dataframe with fields v1 and v2 is generated. Field v1 is the index of starting vertex and v2 is the index of the ending vertex. In Voronoi case a dataframe containing new vertex information is also generated.
| imnetangleslenghts | Count edge angles and lenghts |
| -v <name> | vertex information |
| -e <name> | edge information |
This command is used to calculate angle and distance information from a graph.
| imbuildnet | Build a line graph suitable for vecfld command |
| -v <name> | vertex information |
| -e <name> | edge information |
| -dout <name> | line graph |
| -w <width> | image width |
| -h <height> | image height |
This command is used to quickly build a linegraph for vecfld.
Example: In this example we use a point field to create Delaunay and Voronoi diagrams.
... # create delaunay graph imvoronoi -d points -dout delaunay -t imnetangleslenghts -v points -e delaunay imbuildnet -v points -e delaunay -dout delaunaynet -w 512 -h 512 # create a voronoi graph imvoronoi -d points -dout voronoi -dout2 voronoipoints imnetangleslenghts -v voronoipoints -e voronoi imbuildnet -v points -e delaunay -dout delaunaynet -w 512 -h 512 # show graphs over the bitmap mkgrp g bgimg g -img oldimage vecfld g -n delaunay -d delaunaynet -co blue vecfld g -n voronoi -d voronoinet -co green show g
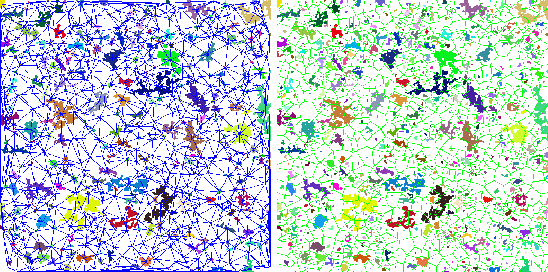
| imfractal | Count fractal features from an image |
| -i <name> | source image name |
| -dout <name> | feature information |
| [-curve <name>] | curve points where features are extracted |
| -xadd <size> | horizontal step |
| -yadd <size> | vertical step |
| -xsize <size> | horizontal max.size |
| -ysize <size> | vertical max. size |
This command calculates a few fractal features from a gray image.
Example: In this example we calculate fractal features from an image.
imload -n image -f paper.gif
imfractal -i image -dout features -curve curve -xadd 512 -yadd 512 \
-xsize 512 -ysize 512 -pnts 7
mkgrp g
axis g -ax x -f curve.x
datplc g -n plt -ax x -f curve.x -co blue
axis g -ax y -f curve.y
datplc g -n plt -ax y -f curve.y -co blue
viewp g -dz -111
show g